What is AWS HealthOmics?
AWS HealthOmics is a HIPAA-eligible service that accelerates clinical diagnostic testing, drug discovery, and agriculture research by fully managing the complex infrastructure behind your bioinformatics workflows. HealthOmics supports industry-standard workflow languages (WDL, Nextflow, CWL) and seamlessly scales bioinformatics infrastructure to support data from tens of thousands of tests per day — all with predictable cost per-sample. HealthOmics handles the technical complexities like managing compute resources and maintaining workflow engines so you can focus entirely on scientific breakthroughs.
Primary use cases include:
- Clinical Diagnostics
- Drug Discovery
- Agricultural Research
Key benefits are:
- Scale without complexity
- Focus on science, not infrastructure
- Built-in compliance features
Pre-requisites for running the bioinformatics workflow on AWS HealthOmics.
There are a total of three prerequisites for deploying the workflow on AWS HealthOmics:
- Create an S3 bucket to store reads and to collect the outputs.
- Create an ECR repo to hold the Docker images of the tools that would be used in the workflow.
- Create an IAM service role for HealthOmics that would allow it to access S3 and ECR.
You can refer to this demo Nextflow pipeline or you can use your own.
How to deploy a Nextflow Pipeline on AWS HealthOmics?
Step 1: Go to the HealthOmics homepage on AWS
Search for HealthOmics in the search bar and navigate to the Private workflows section on the left-hand side.
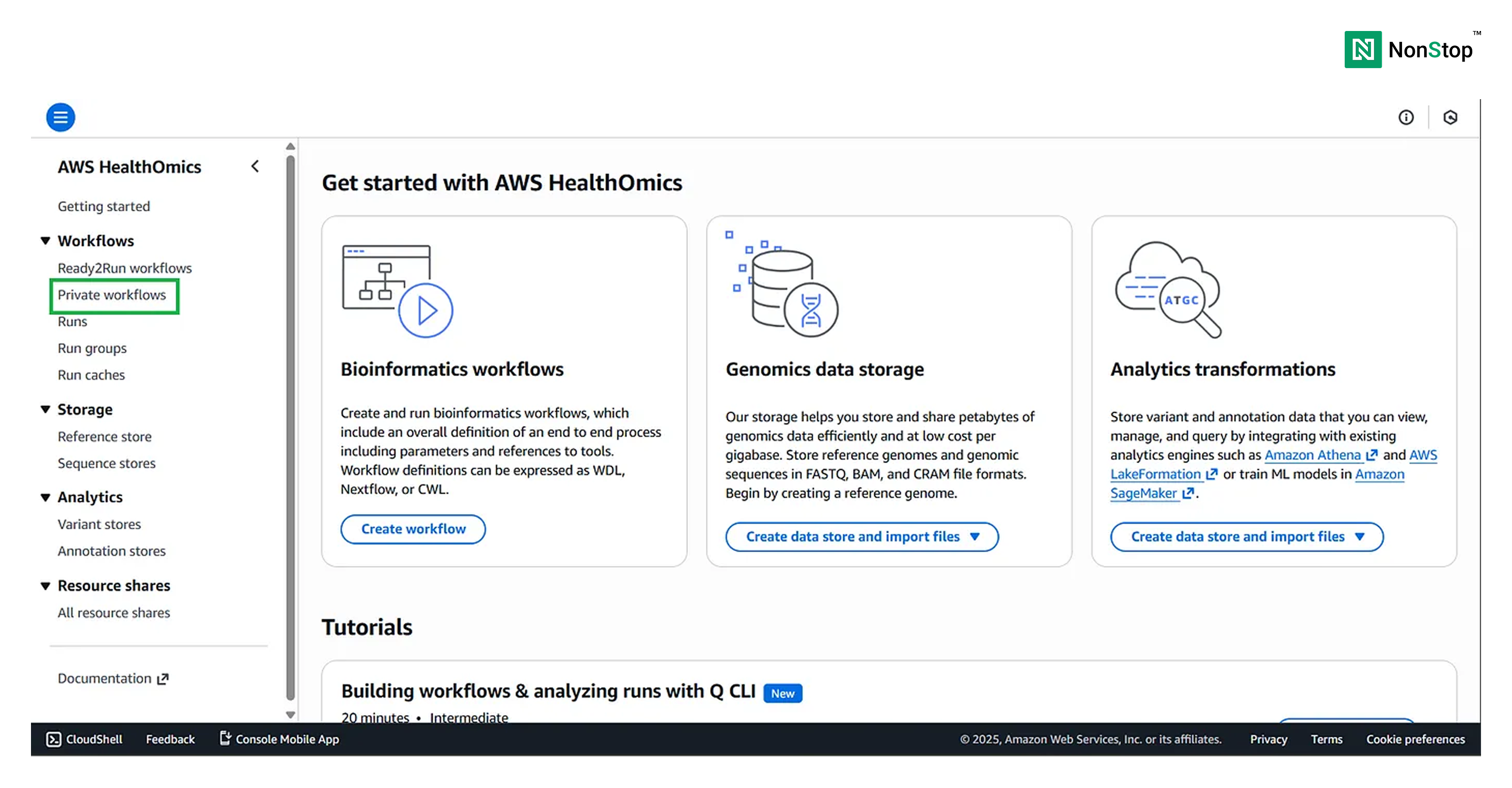
Note: AWS HealthOmics also provides Ready2Run workflows, which you can run directly on your data. The costing for each of those workflows is provided along with them.
Also, under the Storage section, you can see: Reference Stores and Sequence Stores. You can use these stores (i.e., databases) to store your sequences and references. However, for this tutorial, we will stick to S3 buckets.
Step 2: Create a workflow
Scroll down and simply click on the Create Workflow button.
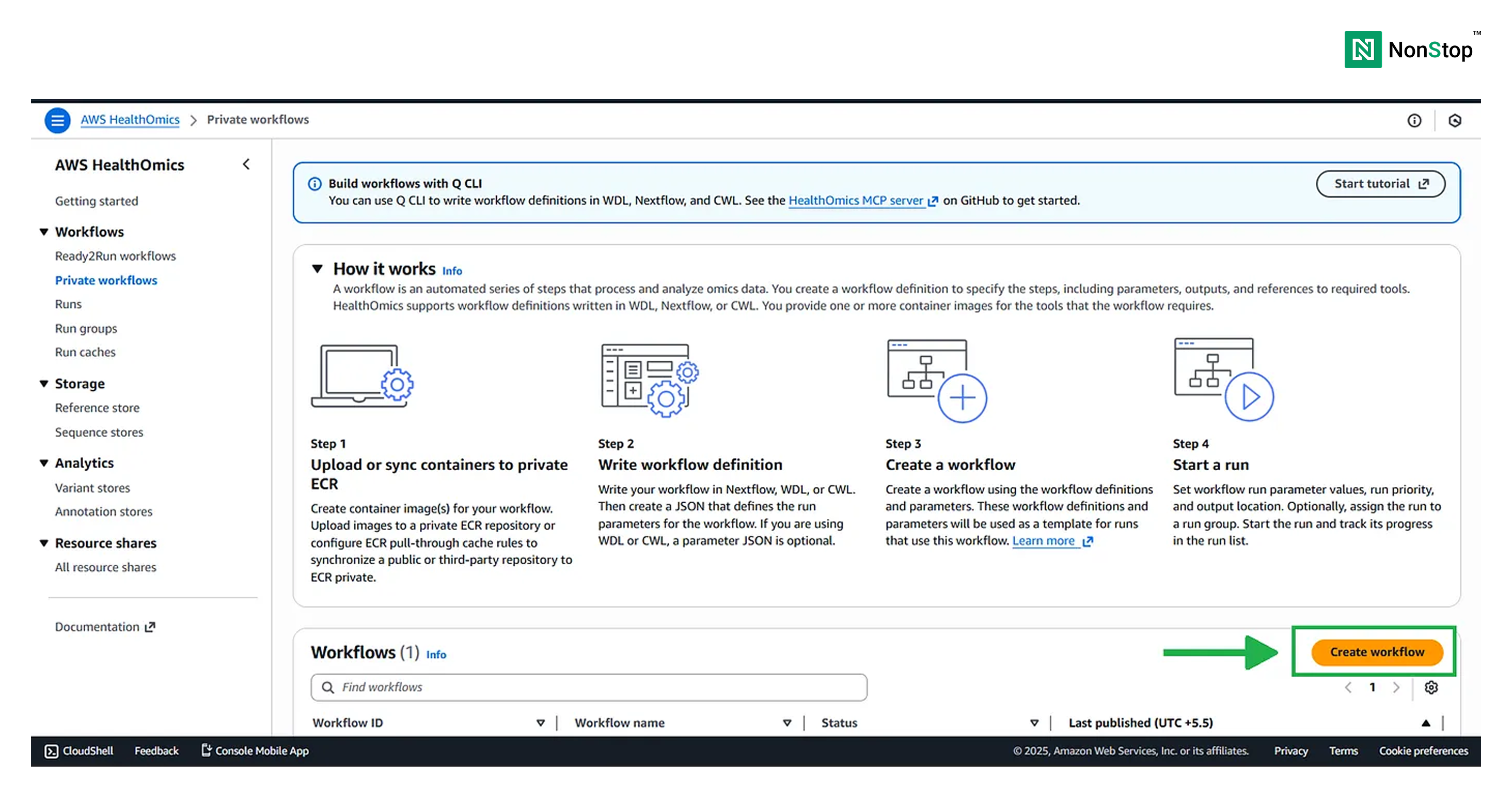
Step 3: Creating the workflow
Creating a bioinformatics workflow in AWS HealthOmics is a straightforward four-step process.
Note: All the fields marked as optional can be skipped. They are as follows:
Step I: Define the workflow
Provide a name for your workflow and a description (optional).
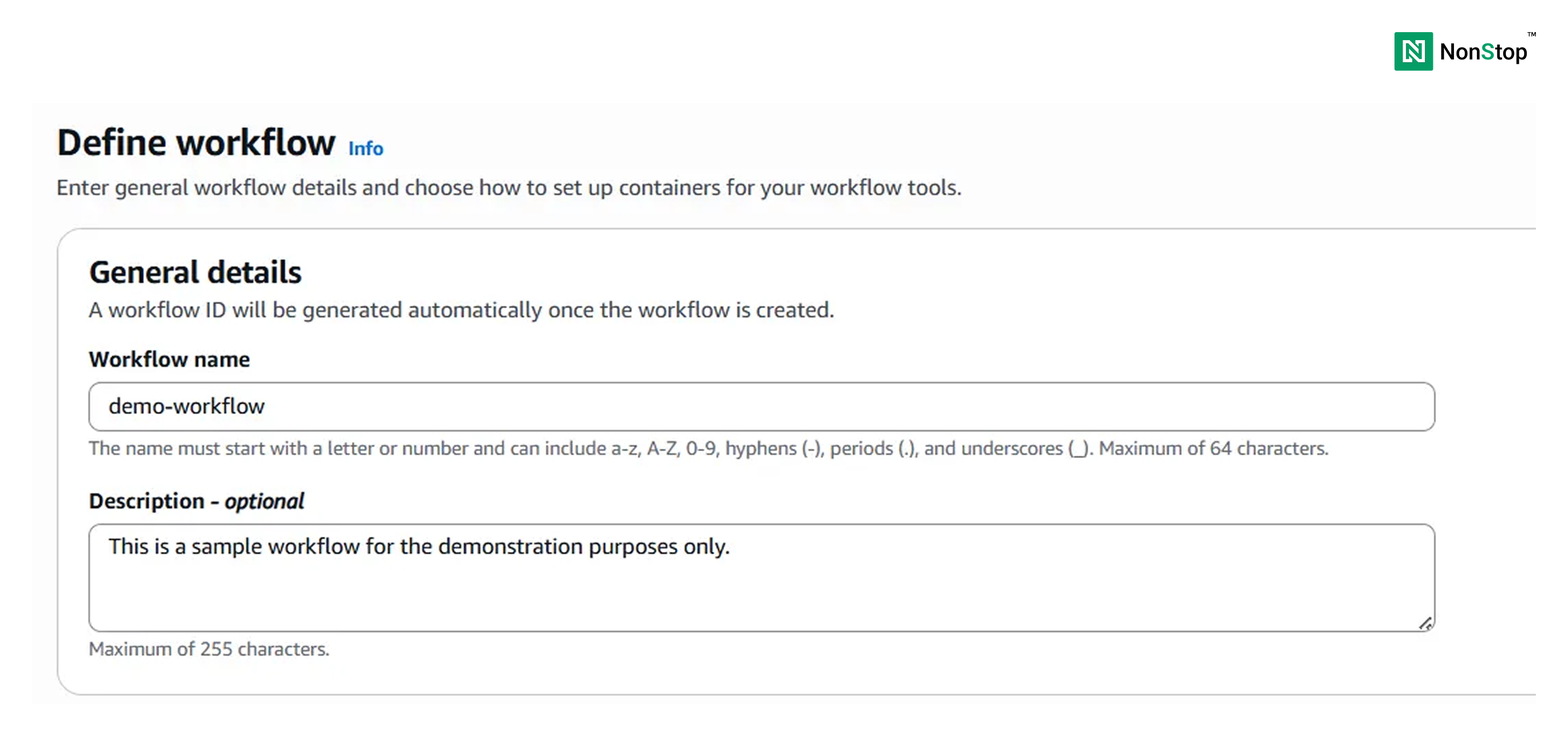
Next, choose the Workflow language. Although this is optional (as HealthOmics would automatically identify it), it is better to select it to reduce latency.
Then, we have the option to select the workflow definition source. It can be any one of the following: S3, a git repository, or direct upload from local source. We shall proceed forward with S3 as all of our data is stored in the S3 bucket.
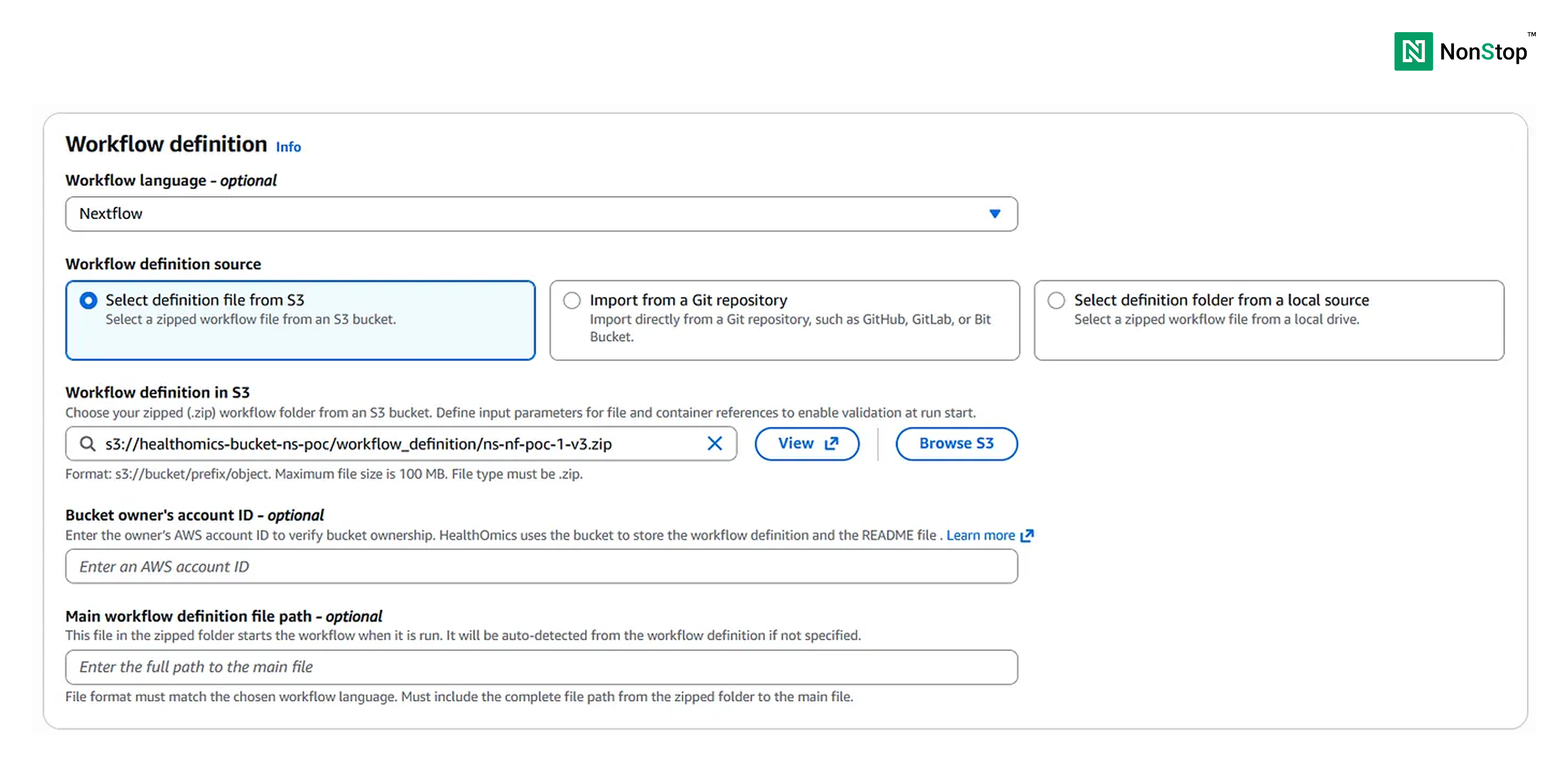
Next, we must select dynamic storage as we don’t have a very high resource requirement.
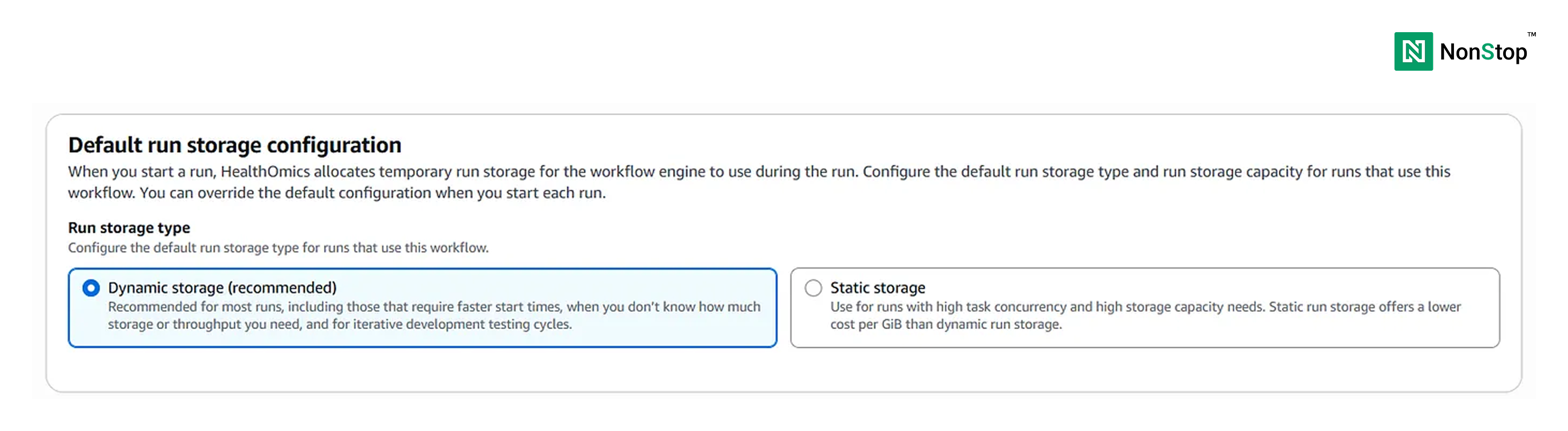
We will leave all the other optional fields blank.
The first step towards building a bioinformatics workflow in AWS HealthOmics is complete!
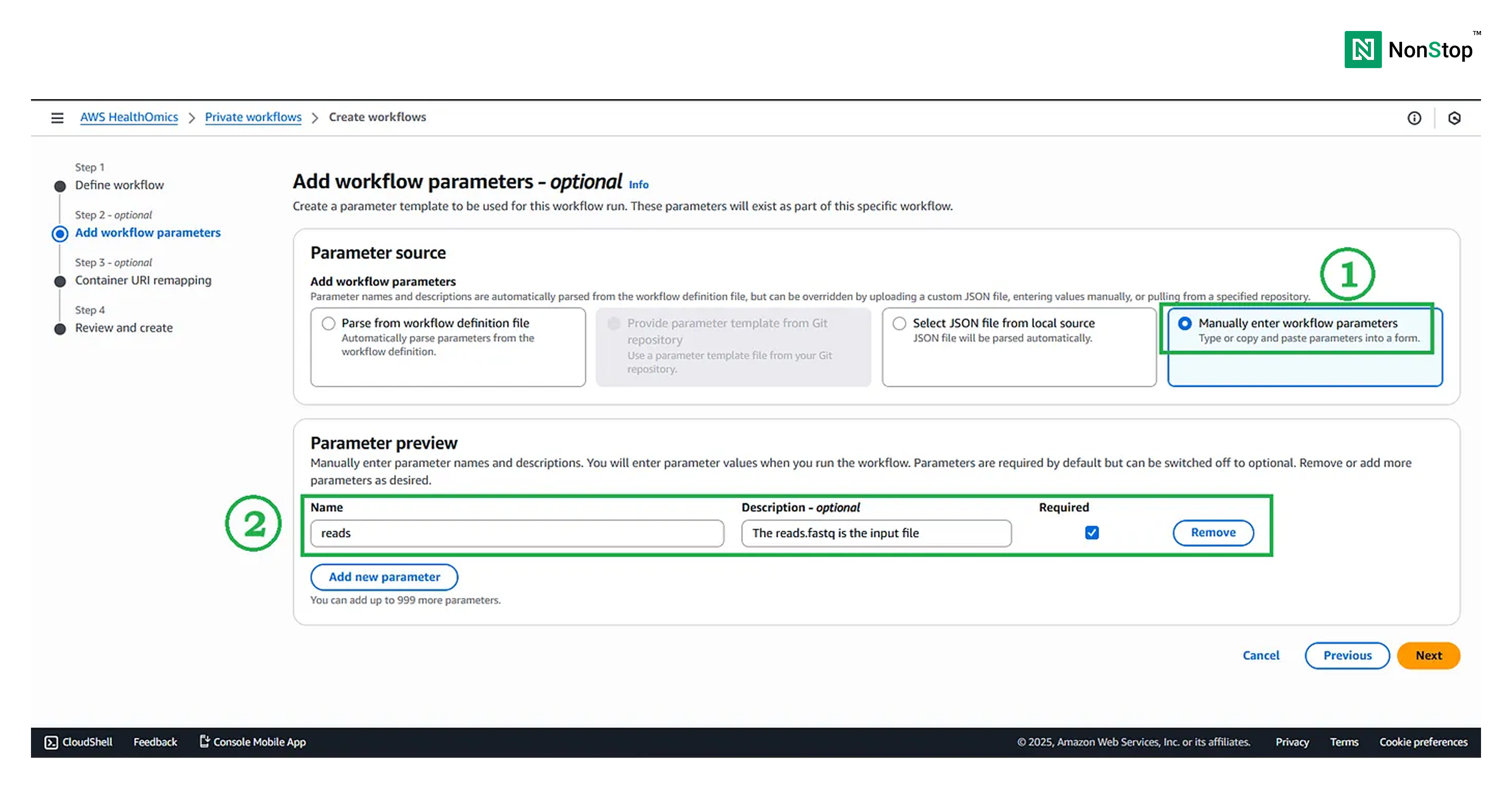
Step II: Add workflow parameters — optional
Although this is an optional step, we shall pass the path to the reads (on S3) via parameters, as our pipeline expects a reads parameter that contains the location of the input reads.
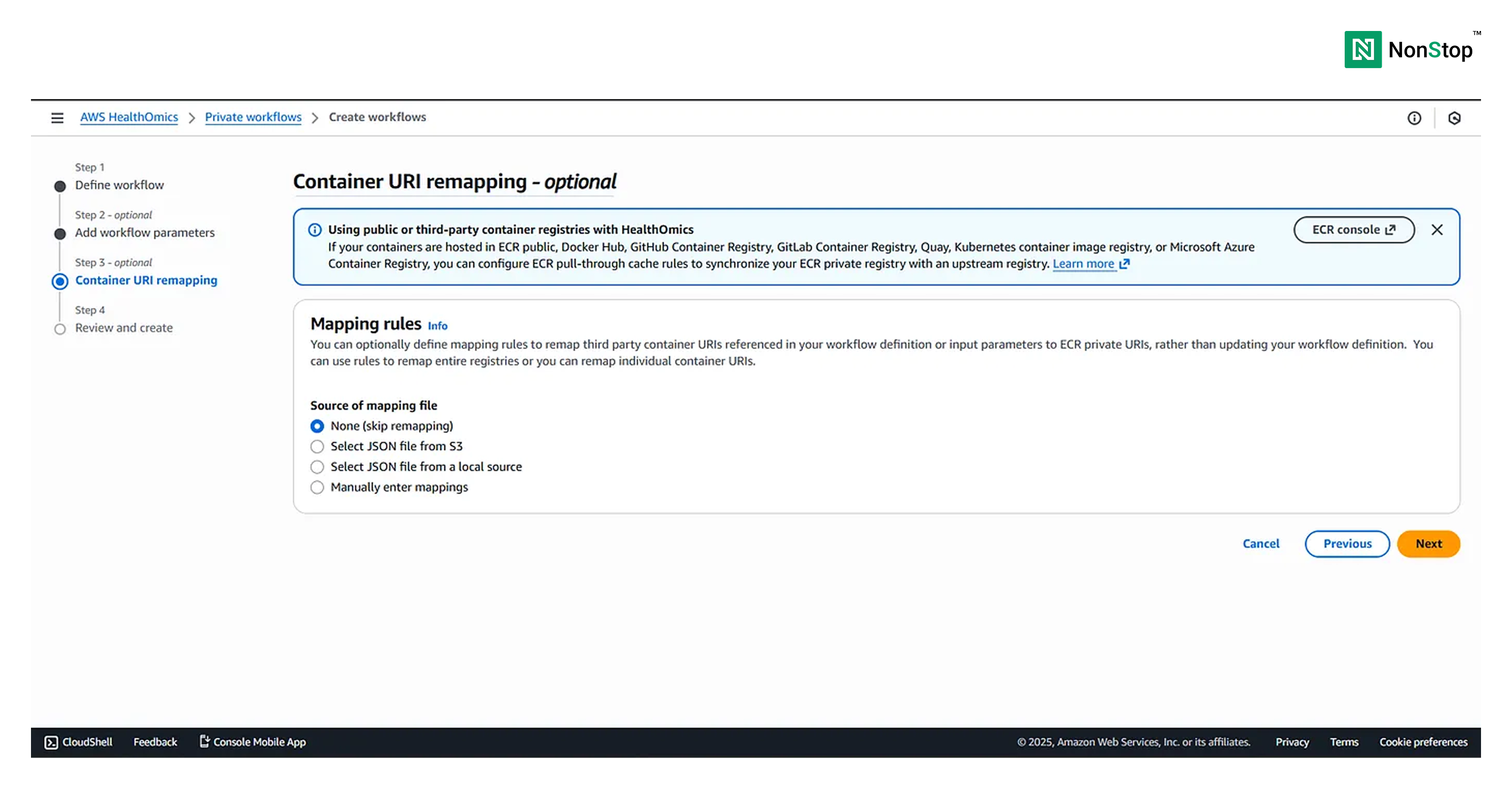
Step III: Container URI remapping — optional
We will choose the source of the mapping file as None (skip remapping). This is because we have already uploaded the Docker images of the tools used to our ECR repo, and we have embedded those links into the source code itself.
Step IV: Review and create
This is just an overview of our configuration. You can edit the fields here and cross-check if anything seems out of place. We are all set to go; you can just scroll to the bottom of the page and click on the Create workflow button.
Congratulations, you just created (probably your first) Nextflow workflow on AWS HealthOmics 🎉 .
HealthOmics might take a few minutes to get everything up and running, so don’t panic if you see such screens for a few minutes.
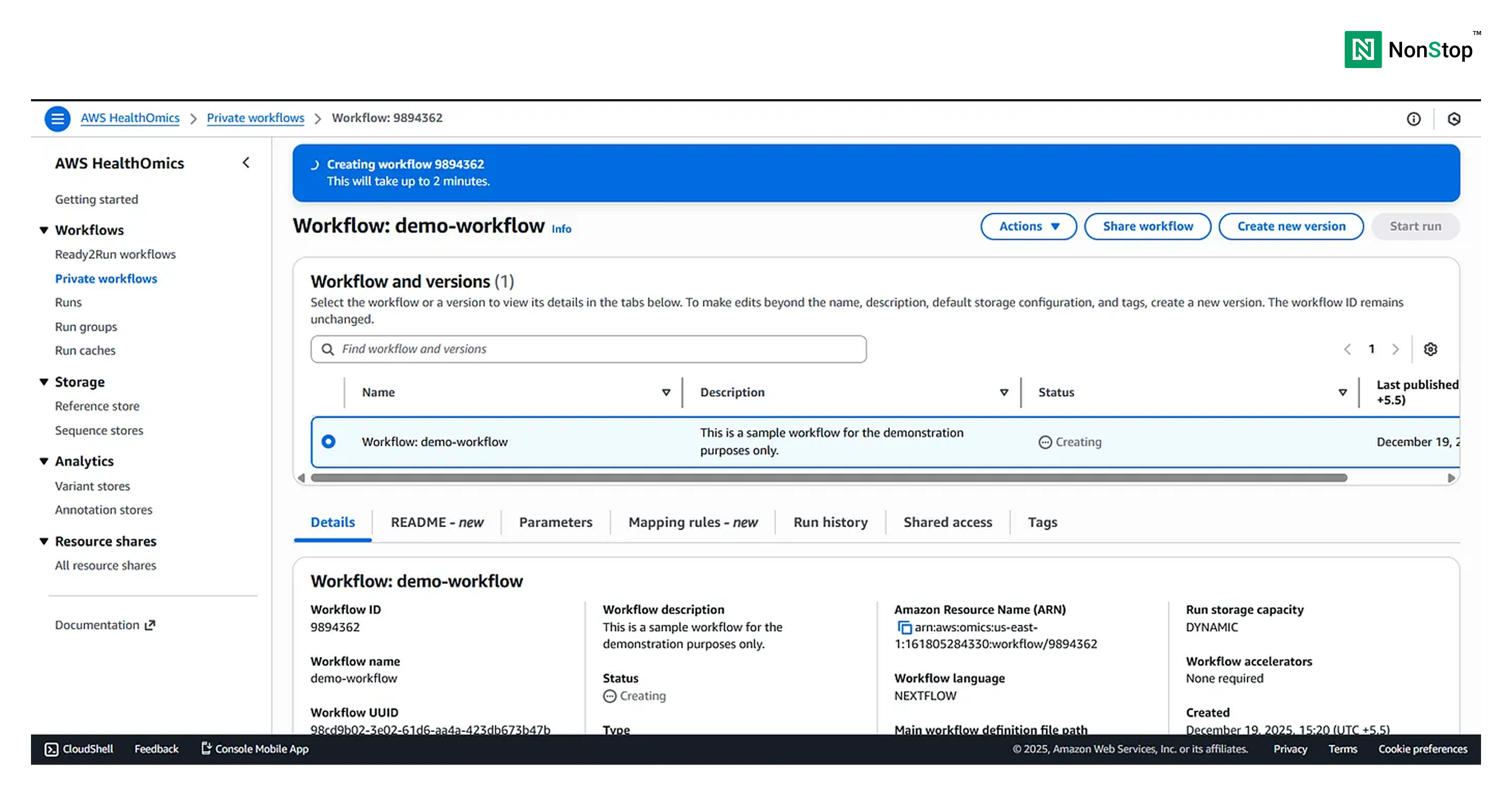
Once the workflow is ready to run, HealthOmics will display a pop-up:
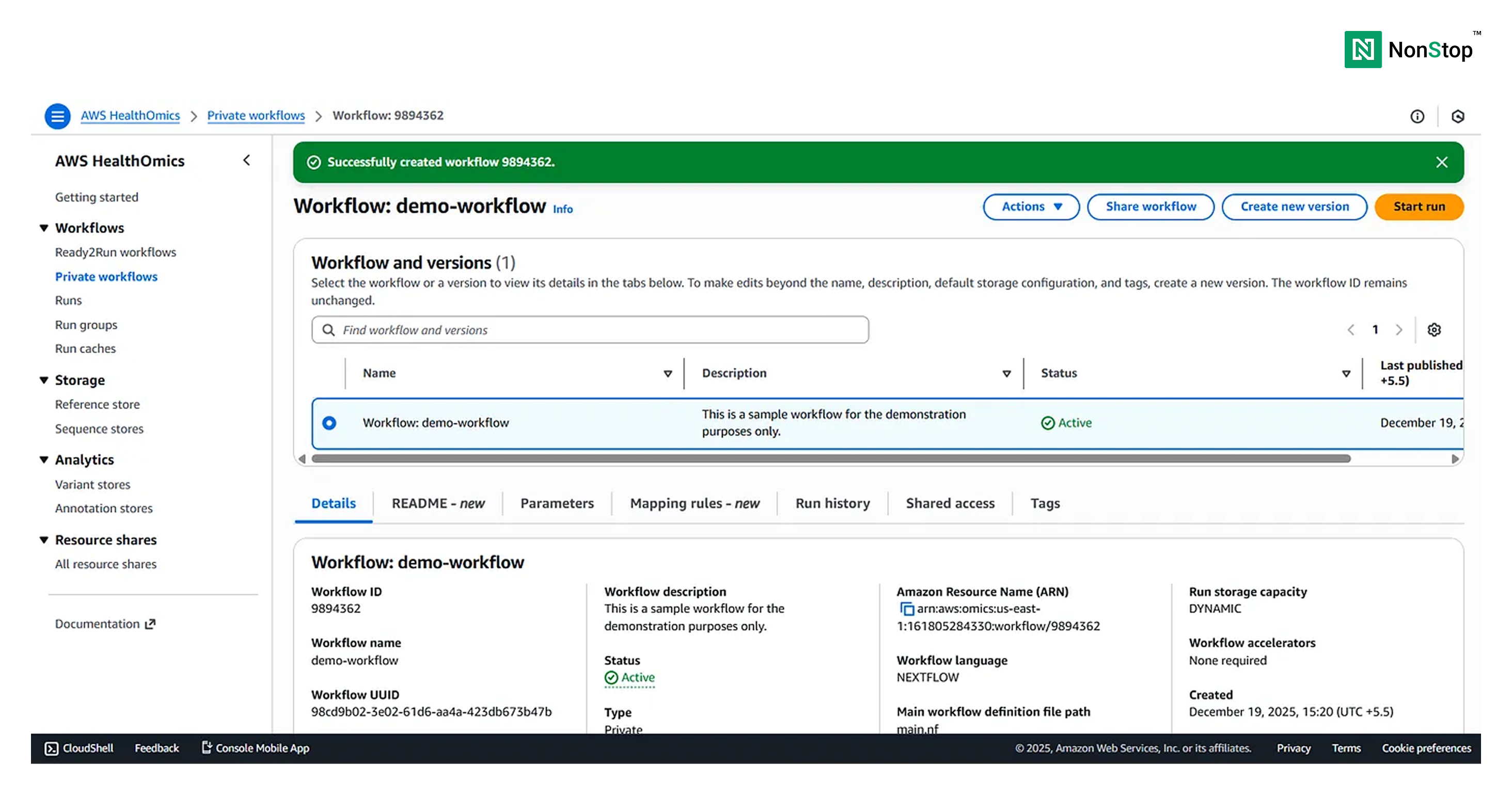
Step 4: Running the workflow
Once your workflow is ready, you can run it with just a single click!
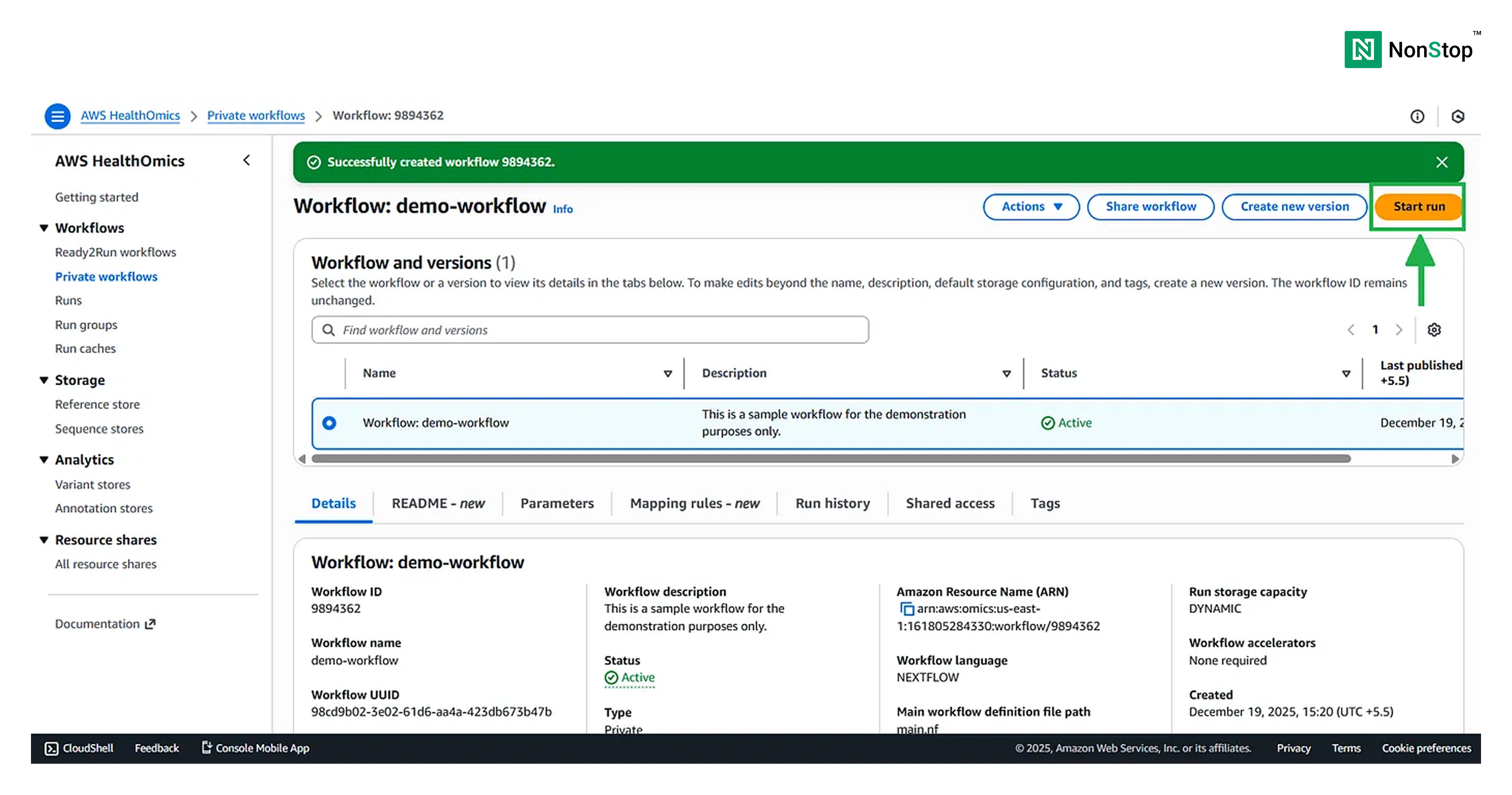
Now, running the workflow is again divided into four steps.
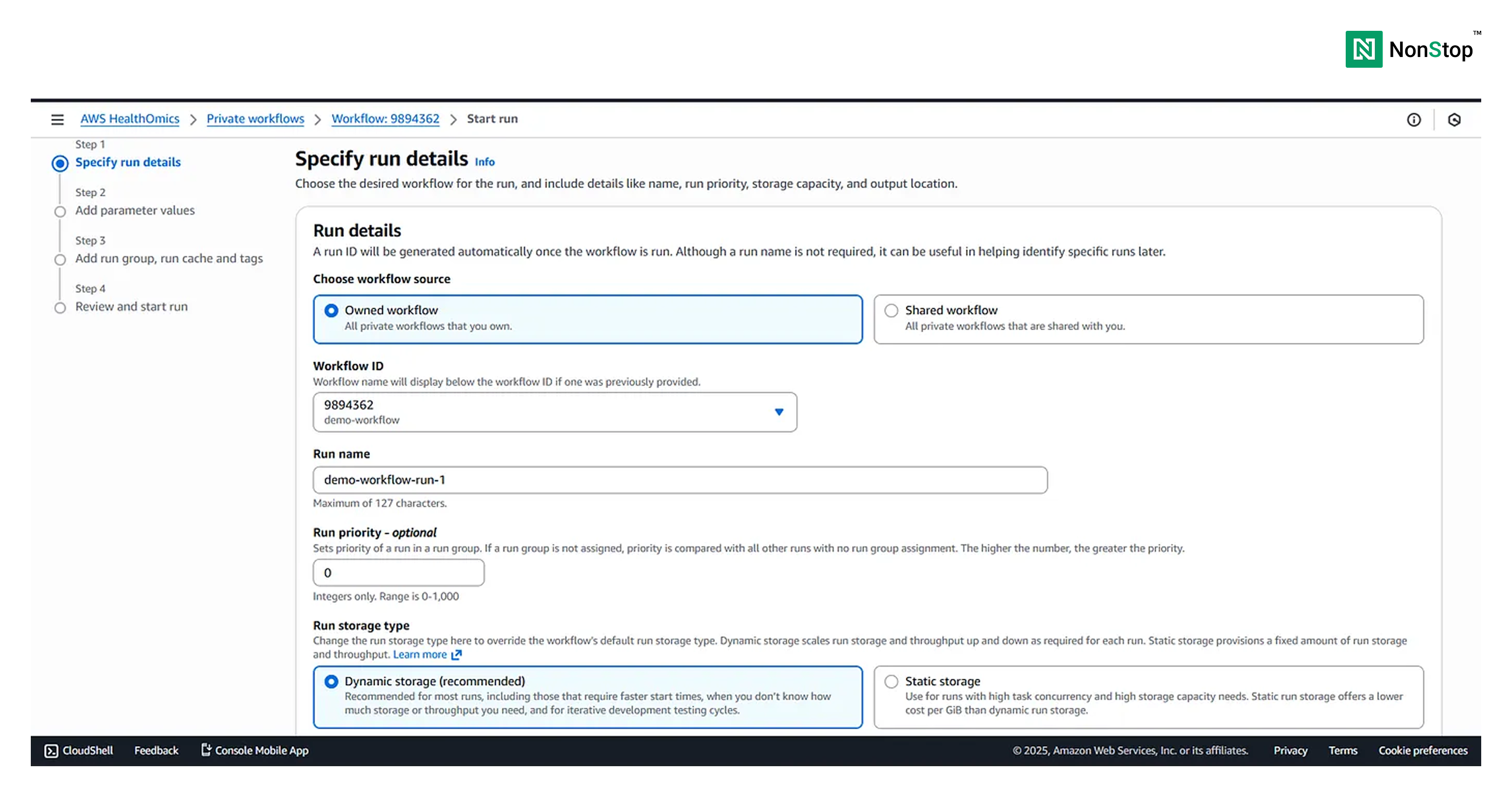
Step I: Specify run details
We need to select the Owned workflow as we just created one. You can provide any name to this particular run. Also, you can provide a run priority; we would be omitting that for now. Lastly, and once again, we choose the run storage type as Dynamic, as our workflow isn’t very resource-heavy.
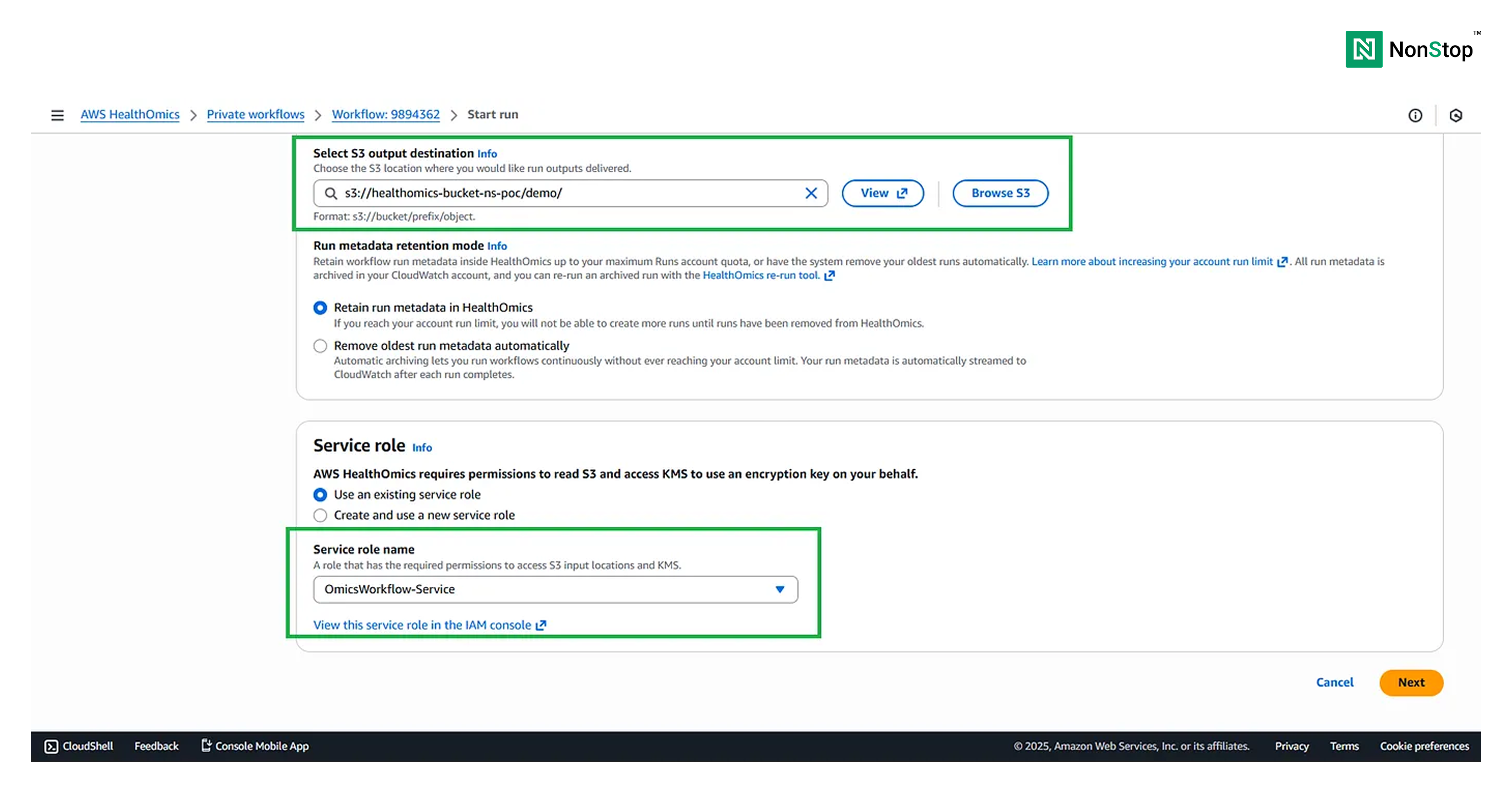
Then, we need to provide the output destination for our workflow, which would again be an S3 bucket.
Also, we need to create an IAM Service which would allow access of S3 to HealthOmics.
Step II: Add parameter values
Next, we would need to provide the values for the parameters we just set. We will choose the manual option as we have only one parameter. We need to provide the path to our input file here (i.e., the reads. fastq file)

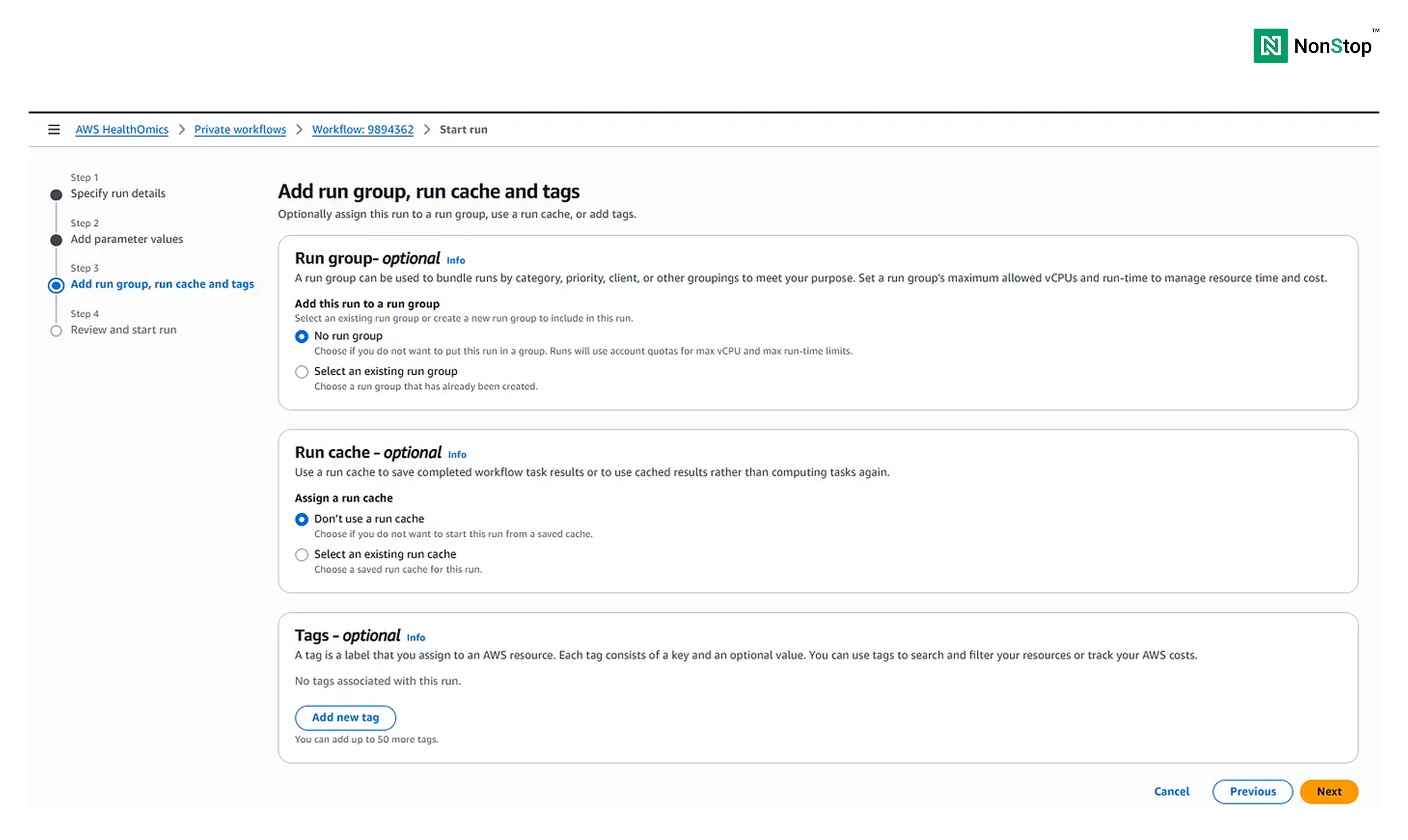
Step III: Add run group, run cache, and tags
This is a completely optional step; it is meant to help organize and optimize industry-grade workflows. We won’t trouble ourselves with this now.
Step IV: Review and start running
Once again, we have the option to edit and cross-check our run configuration. We can safely proceed with the start run.
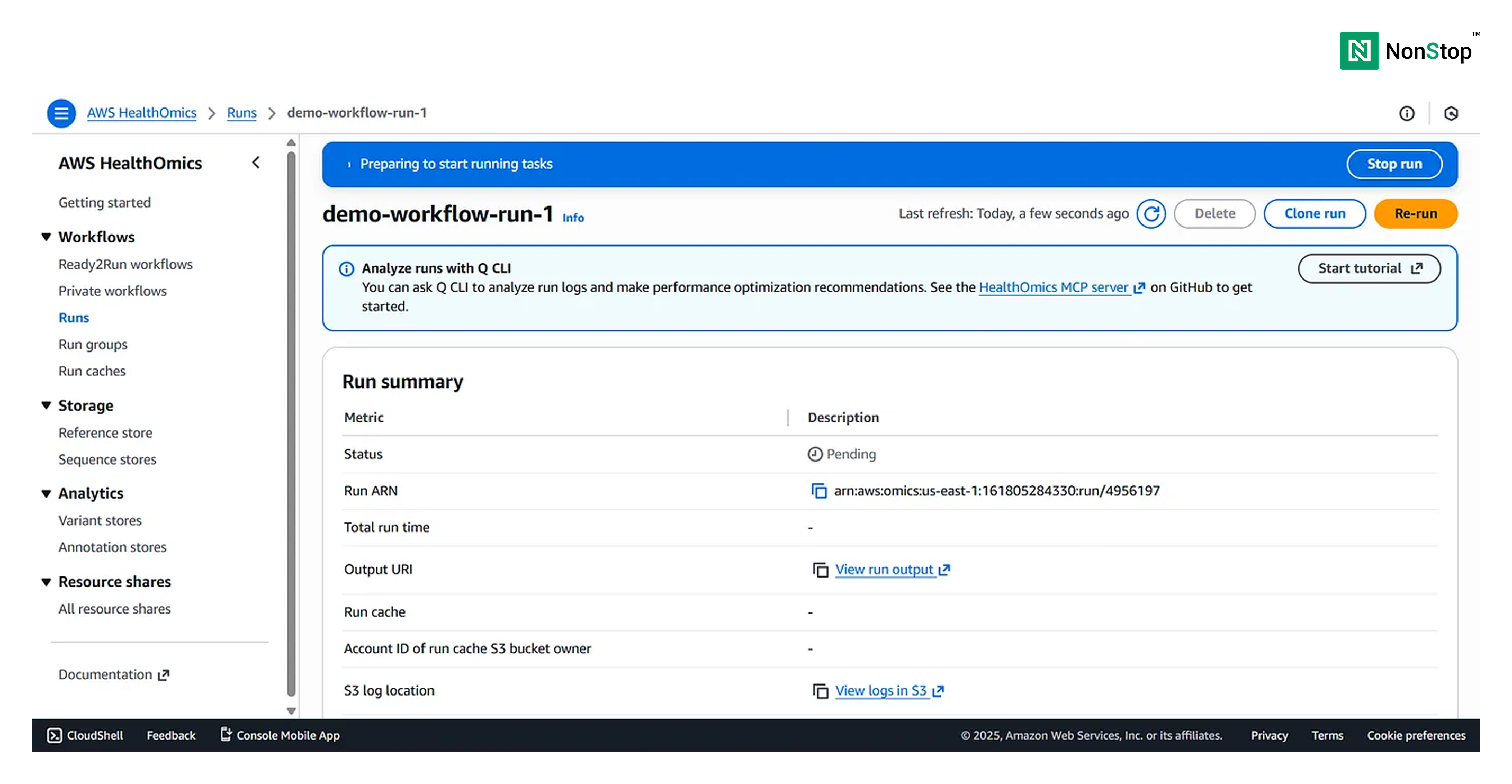
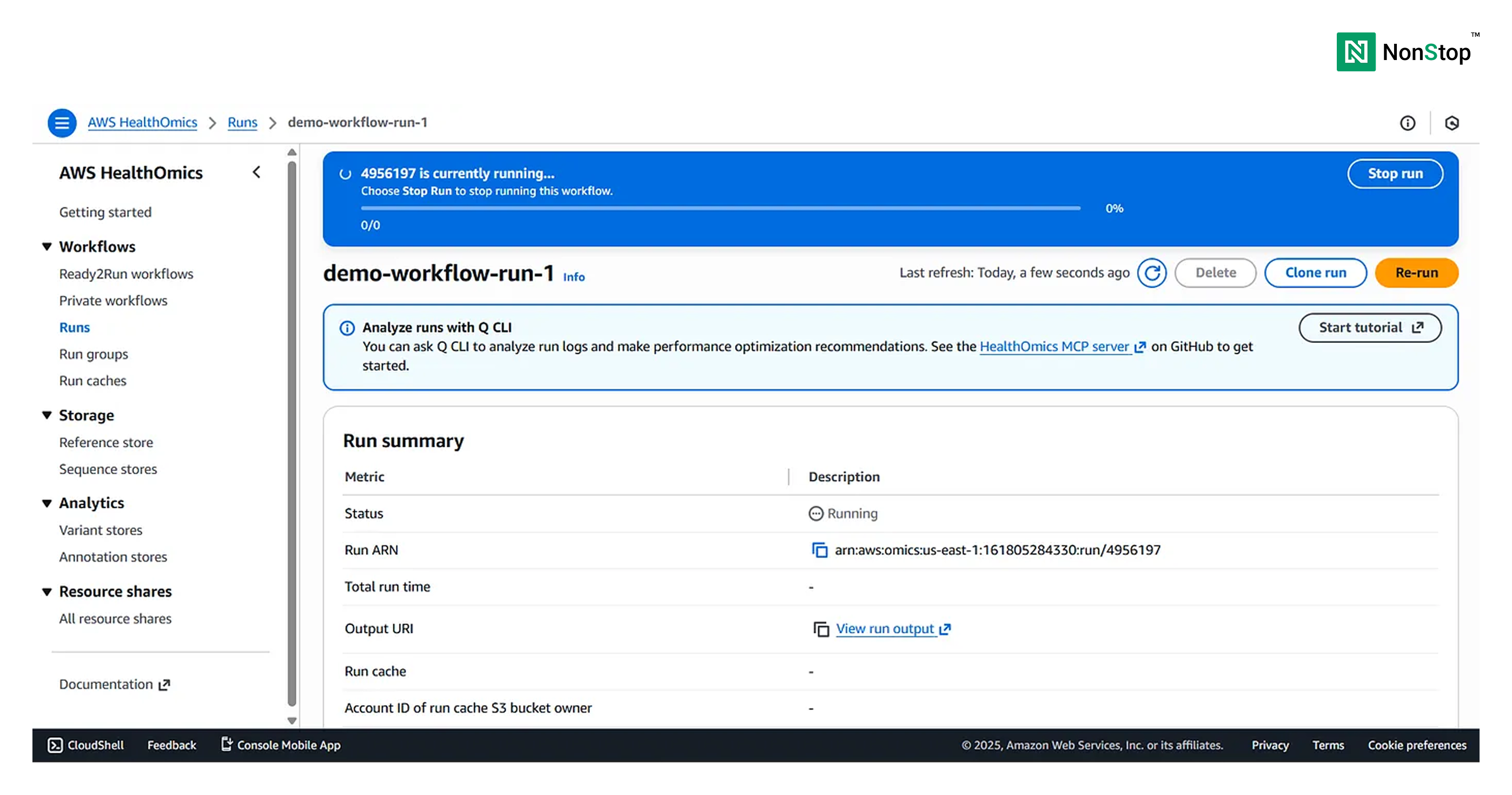
Once the run is successful, you’ll receive the following pop-up:
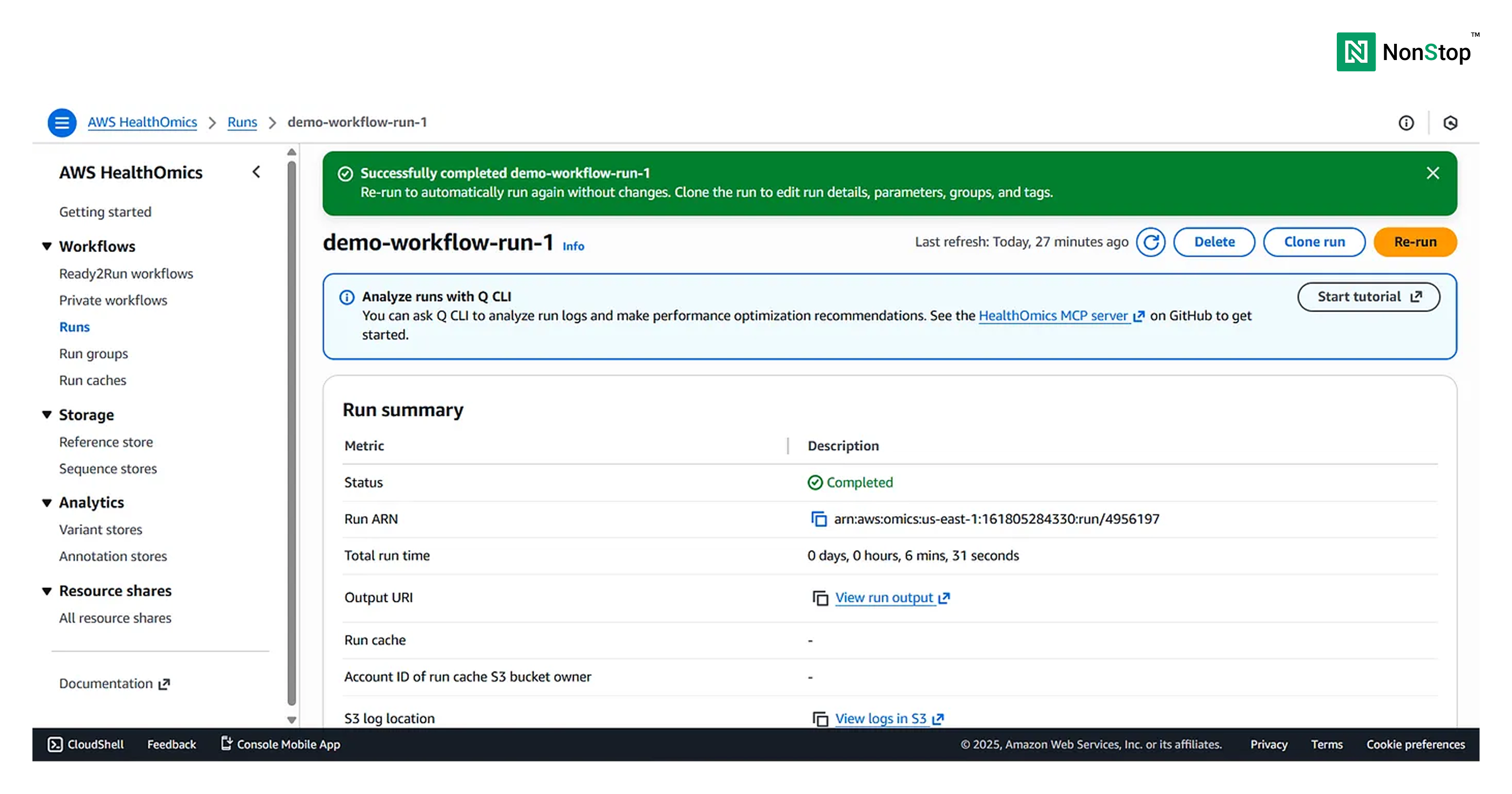
You can view the Run Summary at the bottom of the page:
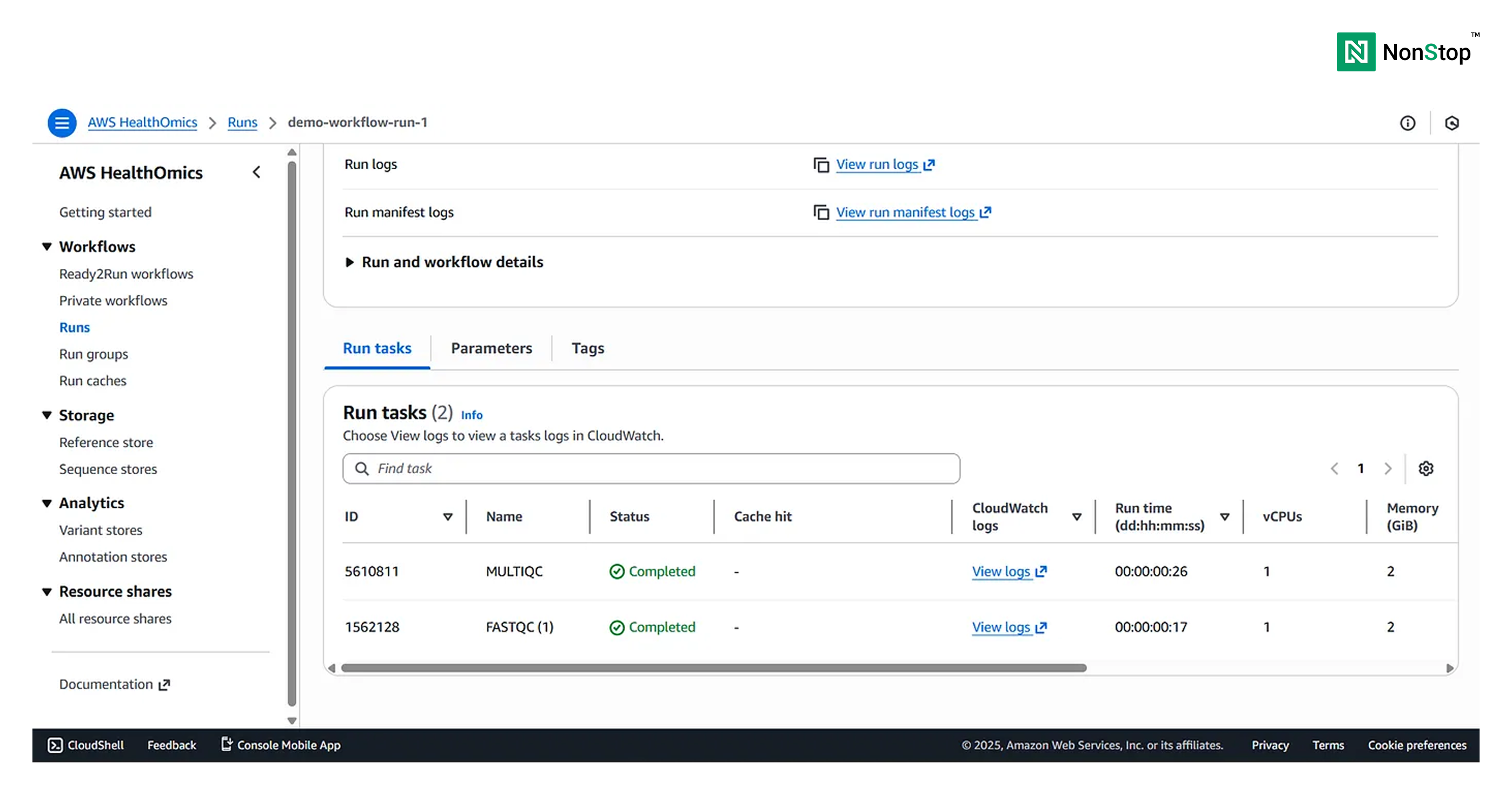
Congratulations 🥳 Your workflow run is complete and successful, the outputs will be present in the S3 bucket (destination provided earlier during Run Configuration).



