When your organization is ready to build or modernize genomics software, whether it's a cloud-native LIMS, a variant interpretation platform, or an EHR-integrated clinical genomics workflow, the choice of development partner becomes one of the most consequential decisions you'll make.
Get it right, and you accelerate time-to-market, reduce technical debt, and build systems that scale with regulatory confidence. Get it wrong, and you inherit years of refactoring, compliance gaps, interoperability nightmares, and platforms that can't handle production-scale sequencing data.
This isn't about finding a vendor who can build software. It's about finding a partner who understands the unique intersection of genomics, healthcare infrastructure, regulatory frameworks, and modern cloud architecture and has proven they can deliver in that exact context.
This article walks through the 12 non-negotiables you should evaluate when choosing a genomics software development partner. These aren't generic software development criteria. They're specific to the complexities of genomics workflows, clinical data integration, compliance requirements, and the technical challenges that emerge when building at the intersection of biology and software.
Why Choosing the Wrong Partner Costs More Than Money
Before diving into the criteria, it's worth understanding what's actually at stake.
In traditional software development, a bad vendor choice might mean delays, budget overruns, or technical debt. In genomics and healthcare, the consequences compound:
- Regulatory non-compliance: A partner unfamiliar with HIPAA,or FDA guidance around software as a medical device (SaMD) can deliver software that cannot legally be deployed in clinical settings. Retrofitting compliance is extraordinarily expensive and sometimes impossible without a complete rebuild.
- Interoperability failure: Genomics software doesn't exist in isolation. It must integrate with EHRs, LIMS, variant databases, clinical decision support systems, and reference genome pipelines. A partner without deep experience in HL7 FHIR, SMART on FHIR, or genomics-specific data standards (like VCF, GVCF, BAM/CRAM formats) will create silos, not solutions.
- Inability to scale: Sequencing throughput is increasing exponentially. Whole genome sequencing generates 100+ GB per sample. If your partner doesn't architect for cloud-native compute, distributed storage, and parallelized bioinformatics pipelines, your platform will collapse under production load.
- Lock-in and technical debt: Many development shops build monolithic, tightly-coupled systems that become unmaintainable. You end up locked into their code, unable to iterate, extend, or bring development in-house without starting over.
The right partner doesn't just deliver software. They provide systems that are compliant, scalable, maintainable, and interoperable by design.
The 12 Non-Negotiables
1. Proven Domain Expertise in Genomics and Clinical Workflows
Why it matters:
Genomics software development is not general-purpose software engineering. The domain complexity is extraordinary: understanding NGS workflows, variant calling pipelines, annotation databases, clinical interpretation frameworks (ACMG/AMP guidelines), and lab reporting standards requires years of hands-on experience.
A partner without this expertise will:
- Misunderstand your requirements
- Propose architectures that don't align with genomics data patterns
- Underestimate the computational and storage requirements
- Build systems that bioinformaticians and lab directors find unusable
What to look for:
- Portfolio of genomics or precision medicine projects (LIMS, variant interpretation platforms, clinical reporting tools, population genomics systems)
- Team members with bioinformatics backgrounds or deep genomics product experience
- Familiarity with standard tools and formats: GATK, bcftools, VCF/GVCF, FASTQ, BAM/CRAM, ClinVar, gnomAD, HGVS nomenclature
- Understanding of clinical genomics workflows: sample accessioning, QC, alignment, variant calling, annotation, interpretation, reporting
Questions to ask:
- Can you walk me through a recent genomics project you've built? What was the pipeline architecture?
- How do you handle VCF normalization and annotation at scale?
- What's your experience with ACMG/AMP variant classification frameworks?
If they can't speak fluently about these topics, they're not domain-ready.
2. Deep Understanding of Healthcare Regulatory Requirements
Why it matters:
Genomics software operating in clinical settings is subject to HIPAA, potentially FDA oversight (if it provides clinical decision support or diagnostic outputs), regulations (if integrated with lab workflows), and state-specific genetic privacy laws.
A partner who treats compliance as an afterthought will deliver software that:
- Exposes PHI without proper access controls or audit logging
- Lacks the documentation required for regulatory submissions
- Cannot pass SOC 2 or HITRUST audits required by health systems
- Creates liability for your organization
What to look for:
- Direct experience building HIPAA-compliant systems with technical safeguards (encryption at rest/in transit, role-based access control, audit logs, BAAs)
- Understanding of FDA guidance on SaMD and clinical decision support
- Experience architecting systems for SOC 2 Type II or HITRUST certification
- Knowledge of GDPR considerations for genomics data (right to deletion is complex with genomic datasets)
Questions to ask:
- How do you architect for HIPAA compliance from day one?
- What's your approach to audit logging and access controls for PHI?
If they say we'll add compliance later, walk away.
3. Technical Architecture Built for Scale and Performance
Why it matters:
Genomics data is massive. A single whole genome generates 100-200 GB of raw data. Clinical labs processing hundreds or thousands of samples per month generate petabytes annually.
Your partner must architect for:
- Distributed storage: Object storage (S3, GCS, Azure Blob) with lifecycle policies
- Parallelized compute: Container orchestration (Kubernetes), workflow engines (Nextflow, Cromwell, AWS Batch), autoscaling
- Efficient data formats: CRAM for storage compression, indexed VCF for query performance
- Streaming and async processing: Queue-based architectures to decouple upload, processing, and reporting
What to look for:
- Experience with cloud-native architectures (AWS, GCP, Azure)
- Proficiency with bioinformatics workflow orchestration (Nextflow, Snakemake, Cromwell/WDL)
- Understanding of computational genomics requirements (CPU/memory per sample, parallelization strategies)
- Use of modern data engineering practices: event-driven architecture, microservices where appropriate, serverless functions for lightweight tasks
Architecture red flags:
- Proposing monolithic architectures for multi-step genomics pipelines
- No plan for handling file sizes exceeding RAM
- Suggesting on-premise infrastructure without a hybrid cloud strategy
- Lack of autoscaling or load management strategy
Questions to ask:
- How would you architect a variant calling pipeline for 500 WGS samples per month?
- What's your approach to managing compute costs while maintaining fast turnaround times?
- How do you handle pipeline failures and reprocessing?
4. EHR and LIMS Integration Expertise
Why it matters:
Genomics software rarely operates in isolation. Clinical genomics workflows require:
- EHR integration: Pulling patient demographics, clinical history, and indication for testing; pushing genomic results back into the EHR for clinician review
- LIMS integration: Tracking sample lifecycle, accessioning, QC status, batch processing
- Variant database integration: ClinVar, gnomAD, COSMIC, dbSNP for annotation and interpretation
A partner without interoperability experience will:
- Build siloed systems that require manual data entry
- Fail to implement HL7 v2, FHIR, or SMART on FHIR standards
- Create brittle point-to-point integrations that break with vendor updates
What to look for:
- Demonstrated experience with HL7 FHIR (especially Genomics Reporting IG)
- SMART on FHIR app development for EHR-integrated clinical decision support
- Integration with commercial LIMS platforms (LabWare, Thermo Fisher, Illumina BaseSpace)
- API-first design with RESTful or GraphQL interfaces
- Experience with healthcare data exchange standards (HL7 v2, CDA, FHIR)
Questions to ask:
- How do you handle EHR integration? What standards and frameworks do you use?
- Can you build a SMART on FHIR app that launches from within Epic or Cerner?
- What's your experience integrating with commercial LIMS platforms?
If they've only built standalone tools, interoperability will be a painful learning curve.
5. Security-First Development Practices
Why it matters:
Genomic data is among the most sensitive data types. It's immutable, identifiable, and reveals information about biological relatives. A breach has lifetime consequences for patients.
Your partner must embed security throughout the development lifecycle:
- Threat modeling during design
- Secure coding practices (OWASP Top 10)
- Encryption at rest and in transit
- Multi-factor authentication, SSO integration (SAML, OAuth)
- Regular penetration testing and vulnerability scanning
- Incident response planning
What to look for:
- Security certifications or training (CISSP, CEH, or equivalent)
- Use of infrastructure-as-code with security policies (Terraform, CloudFormation, with tools like Checkov)
- Container security scanning (Snyk, Aqua, Trivy)
- Secrets management (AWS Secrets Manager, HashiCorp Vault)
- Regular third-party security audits
Questions to ask:
- What's your approach to secure software development lifecycle?
- How do you manage secrets and credentials?
- Do you conduct regular penetration testing?
If security isn't mentioned until you bring it up, it's not part of their culture.
6. Transparent, Predictable Engagement Model
Why it matters:
Genomics projects are complex, and requirements often evolve as you learn. Your partner's engagement model must support:
- Iterative development with feedback loops
- Transparent pricing (fixed price is often a red flag for complex builds)
- Clear ownership of code and IP
- Flexibility to scale the team up or down
What to look for:
- Agile or iterative development methodology
- Regular sprint reviews and demos
- Transparent backlog and roadmap visibility
- Clear IP ownership (you should own the code)
- Ability to transition knowledge to your internal team
Engagement models to consider:

Questions to ask:
- What does your typical engagement look like?
- How do you handle scope changes?
- Who owns the code and IP?
- What does your offboarding process look like if we want to bring development in-house?
7. Experience with Modern DevOps and Infrastructure
Why it matters:
Genomics platforms must be reliable, maintainable, and deployable across environments. Your partner should implement:
- CI/CD pipelines for automated testing and deployment
- Infrastructure as code
- Containerization (Docker) and orchestration (Kubernetes)
- Monitoring, logging, and alerting (CloudWatch, Datadog, ELK stack)
- Disaster recovery and backup strategies
What to look for:
- Use of modern DevOps tooling (GitHub Actions, GitLab CI, Jenkins, CircleCI)
- Infrastructure automation (Terraform, Pulumi, CloudFormation)
- Monitoring and observability culture
- Environment parity (dev/staging/prod)
- Documented runbooks and operational procedures
Questions to ask:
- What does your CI/CD pipeline look like?
- How do you handle infrastructure provisioning?
- What's your approach to monitoring and incident response?
8. Strong QA and Testing Culture
Why it matters:
In clinical genomics, software errors can lead to misdiagnosis. Robust testing is non-negotiable:
- Unit tests for business logic
- Integration tests for pipeline workflows
- End-to-end tests for user workflows
- Performance tests for scale
- Validation against gold-standard datasets
What to look for:
- Test coverage expectations (>80% for critical paths)
- Automated testing in CI/CD
- Use of test frameworks appropriate to the stack
- Experience with bioinformatics validation (comparing pipeline outputs to known truth sets)
Questions to ask:
- What's your testing strategy?
- How do you validate bioinformatics pipelines?
- What test coverage do you typically achieve?
9. Long-Term Maintainability and Documentation
Why it matters:
You're not just buying a v1.0 product. You're building a platform that will evolve for years. Your partner must deliver:
- Clean, readable, well-documented code
- Architectural documentation
- API documentation (OpenAPI/Swagger)
- User guides and admin manuals
- Runbooks for operations
What to look for:
- Code review practices
- Use of linters and formatters
- Documentation as part of the definition of done
- Knowledge transfer sessions
Questions to ask:
- What documentation do you deliver?
- How do you ensure code quality?
- What does knowledge transfer look like?
10. Cultural Fit and Communication
Why it matters:
You'll work closely with this team for months or years. Cultural alignment matters:
- Clear, proactive communication
- Collaborative mindset (partner, not vendor)
- Willingness to challenge assumptions respectfully
- Responsiveness and accountability
What to look for:
- Responsiveness during the evaluation phase
- The quality of the discovery questions they ask
- Transparency about risks and tradeoffs
- References from past clients about communication
Questions to ask past clients:
- How was communication throughout the project?
- Did they raise issues proactively?
- Would you work with them again?
11. Post-Launch Support and Maintenance
Why it matters:
Software doesn't end at launch. You need:
- Bug fixes and patches
- Performance optimization
- Security updates
- Feature enhancements
- SLA-backed support
What to look for:
- Clear support and maintenance offerings
- SLAs with response times
- On-call support for production issues
- Roadmap for future enhancements
Questions to ask:
- What does post-launch support look like?
- What are your SLAs?
- How do you handle production incidents?
12. Financial Stability and References
Why it matters:
You're making a long-term bet. Your partner must be financially stable with a track record of successful delivery.
What to look for:
- Years in business
- Client retention rates
- Case studies in similar domains
- Referenceable clients
Questions to ask:
- Can you share 2-3 references from genomics or healthcare clients?
- Have you had clients transition projects to other teams? Why?
Decision Framework: Evaluating Partners
Use this scorecard to evaluate potential partners:
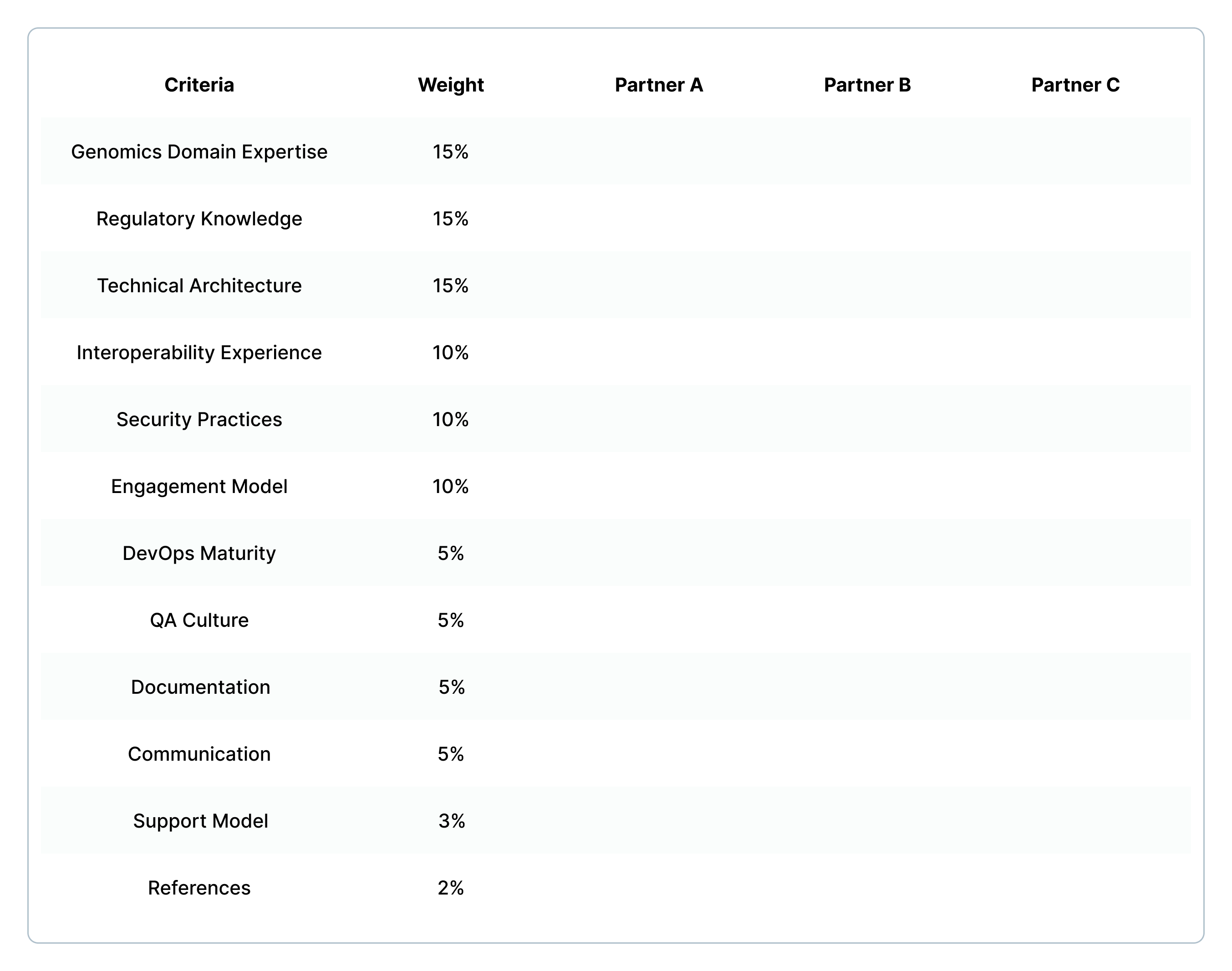
Score each partner 1-5 on each criterion. Multiply by weight - total scores out of 5.
Common Mistakes When Choosing a Partner
Mistake 1: Choosing based on cost alone
The cheapest partner is rarely the best value. Rebuilding after a failed project costs far more than hiring the right team upfront.
Mistake 2: Ignoring domain expertise
General software development skills don't translate directly to genomics. The learning curve is steep and expensive.
Mistake 3: Skipping reference checks
Always speak to past clients. Ask about communication, problem-solving, and post-launch support.
Mistake 4: No technical deep-dive
Don't just meet the sales team. Interview the actual architects and developers who will work on your project.
Mistake 5: Unclear success criteria
Define what success looks like before you start: timelines, quality metrics, performance benchmarks, and compliance requirements.
Choosing a genomics software development partner is not a procurement decision; it's a strategic partnership that shapes your organization's ability to deliver precision medicine, accelerate research, and compete in a rapidly evolving landscape.
The right partner brings more than technical skills. They bring domain wisdom, regulatory foresight, architectural maturity, and a collaborative mindset that makes them an extension of your team.
The 12 non-negotiables outlined in this article provide a framework for making that choice with confidence. Use them to evaluate partners rigorously, ask hard questions, and ultimately select a team that can deliver not just software, but a platform that scales, complies, integrates, and endures.
If your team is exploring modernizing LIMS workflows, building cloud-native genomics tools, or integrating EHR/LIMS systems with AI and compliance built in, NonStop is always open to a conversation. We've spent over a decade helping genomics and healthcare organizations design, engineer, and scale platforms that last.



